Running QM/MM simulations
Running a CP2K standalone job
A standard CP2K build creates multiple executables. It is advised to use the cp2k.psmp executable as this enables the hybrid MPI+OpenMP parallelisation scheme, which can have some performance benefit over using only MPI parallelisation on some machines.
CP2K should be run through a job submission script to run on the compute nodes. The following command will launch CP2K (MPI only), specifing the input and output files, and the number of mpi processes.
export OMP_NUM_THREADS=1
joblauncher (-n procs) cp2k.pmsp -i inputfile.inp -o outputfile.out
The joblauncher will depend on the job launcher on your system, common examples are
mpiexec, srun and aprun.
To run with multiple threads (MPI+OpenMP) the number of threads should be set to a value greater than 1. Typical values where performance may be improved over pure a MPI job are 2, 4, 6, and 8 threads, although this will depend on many things such as your machine architecture, the type of calculation and your system size. As a general rule the number of threads per MPI process has to be chosen so that it evenly divides the number of MPI processes on a node, whilst ensuring that threads sharing memory are in the same NUMA region. The total number of MPI processes will need to be set so that the number of threads per process multiplied by the number of MPI processes gives the total number of cores requested.
Running QM/MM simulations with the GROMACS/CP2K interface
Once all the required input files for the GROMACS/CP2K interface have been created
as described in the GROMACS/CP2K interface QM/MM parameterisation section
you can create the GROMACS tpr file and then launch the MD simulation.
gmx grompp -f sys.mdp -p sys.top -c sys.gro -n sys.ndx -o sys.tpr
joblauncher (n proces) gmx_mpi -s sys.tpr
When generating the tpr file you may get a warning about your system having non-zero
charge. This can safely be ignored for QM/MM calculations by using the -maxwarn option.
The joblauncher will depend on the job launcher on your system, common examples are
mpiexec, srun and aprun
Performance considerations
When running CP2K standalone or together with GROMACS using the GROMACS-CP2K interface, the run times of simulations will be dominated by the QM and QM/MM calculations within CP2K. The performance of QM/MM simulation using GROMACS together with CP2K is roughly equivalent to that of performing the equivalent simulation using CP2K standalone.
For the sake of reference we include here performance results for a number of CP2K QM/MM benchmarks that are part of the Bioexel QM/MM benchmark suite.
Name |
Type |
Total atoms |
QM atoms |
Functional |
QM Cell Size |
Basis set |
Time step |
MD Type |
Periodic |
|---|---|---|---|---|---|---|---|---|---|
MQAE-BLYP |
solute-solvent |
~16,000 |
34 |
BLYP |
14 x 17 x 11 |
DZVP-MOLOPT |
1 fs |
NVE |
Y |
MQAE-B3LYP |
B3LYP |
6-31Gxx |
|||||||
MQAE-B3LYP-large |
28 x 34 x 22 |
||||||||
CBD_PHY-PBE |
phytochrome |
~168,000 |
68 |
PBE |
25 x 25 x 25 |
DZVP-MOLOPT |
|||
CBD_PHY-PBE0 |
PBE0 |
HFX_BASIS TZV2P |
|||||||
ClC-19-BLYP |
ion channel |
~150,000 |
19 |
BLYP |
18 x 18 x 18 |
DVZP-MOLOPT |
|||
ClC-253-BLYP |
253 |
27 x 25 x 25 |
|||||||
GFP_ScaleQM |
fluorescent protein |
~28,000 |
20 |
BLYP |
40 x 40 x 40 |
DZVP-GTH-BLYP |
NVT |
N |
|
32 |
|||||||||
53 |
|||||||||
77 |
The raw data for the performance of these benchmarks is available in the QM/MM Benchmarking Data repository. Their performance is summarised below.
Running on CPUs
The time per MD step (s) and performance (ps/day) on ARCHER2 is reported in the Tables below for the benchmark systems. ARCHER2 has 128 cores per node, comprised of two 64-core AMD EYPC processors. More details are given on the ARCHER2 website The results below use MPI+OpenMP with 4 threads per MPI process which was found to, in general, give the best performance.
MQAE (BLYP) |
MQAE (B3LYP) |
ClC (QM 19) |
ClC (QM 253) |
CBD_PHY (PBE) |
CBD_PHY (PBE0) |
|
|---|---|---|---|---|---|---|
Cores |
Time per MD step (s) |
|||||
32 |
12.86 |
20.94 |
46.83 |
168.16 |
112.83 |
266.25 |
64 |
7.38 |
12.38 |
25.82 |
111.86 |
66.95 |
127.51 |
128 |
4.91 |
7.85 |
15.07 |
75.03 |
38.13 |
69.48 |
256 |
3.55 |
5.13 |
57.79 |
24.89 |
40.21 |
|
512 |
24.93 |
|||||
MQAE (BLYP) |
MQAE (B3LYP) |
ClC (QM 19) |
ClC (QM 253) |
CBD_PHY (PBE) |
CBD_PHY (PBE0) |
|
|---|---|---|---|---|---|---|
Cores |
Performance (ps/day) |
|||||
32 |
6.71 |
4.13 |
1.84 |
0.51 |
0.76 |
0.32 |
64 |
11.71 |
6.98 |
3.35 |
0.77 |
1.29 |
0.68 |
128 |
17.60 |
11.01 |
5.73 |
1.15 |
2.27 |
1.24 |
256 |
24.34 |
16.84 |
1.50 |
3.47 |
2.15 |
|
512 |
3.47 |
|||||
Running on GPUs
The time per MD step (s) and performance (ps/day) on Cirrus GPU nodes is reported in the Tables below for the benchmark systems. The Cirrus GPU nodes contain 4 GPUs per node and 20 CPU cores. The GPUs are Nvidia Volta V100’s Here we assign one MPI process per GPU and 10 OpenMP threads per process to make use of the CPU cores. More details are given in the Cirrus documentation
Using the GPU enabled COSMA library was found to not significantly improve the performance.
MQAE (BLYP) |
MQAE (B3LYP) |
ClC (QM 19) |
ClC (QM 253) |
CBD_PHY (PBE) |
CBD_PHY (PBE0) |
||
|---|---|---|---|---|---|---|---|
Cores |
GPUs |
Time per MD step (s) |
|||||
40 |
4 |
15.43 |
25.55 |
55.73 |
136.09 |
||
80 |
8 |
9.47 |
14.32 |
32.80 |
89.75 |
155.98 |
|
160 |
16 |
6.44 |
8.91 |
20.31 |
63.08 |
38.66 |
70.43 |
320 |
32 |
6.36 |
46.62 |
24.79 |
41.19 |
||
MQAE (BLYP) |
MQAE (B3LYP) |
ClC (QM 19) |
ClC (QM 253) |
CBD_PHY (PBE) |
CBD_PHY (PBE0) |
||
|---|---|---|---|---|---|---|---|
Cores |
GPUs |
Performance (ps/day) |
|||||
40 |
4 |
5.60 |
3.38 |
1.55 |
0.63 |
||
80 |
8 |
9.12 |
6.03 |
2.63 |
0.96 |
0.55 |
|
160 |
16 |
13.42 |
9.70 |
4.25 |
1.37 |
2.23 |
1.23 |
320 |
32 |
13.58 |
1.85 |
3.49 |
2.10 |
||
CPU benchmark results
All results are reported for ARCHER2. MPI+OpenMP is used with 4 threads per process.
MQAE-BLYP
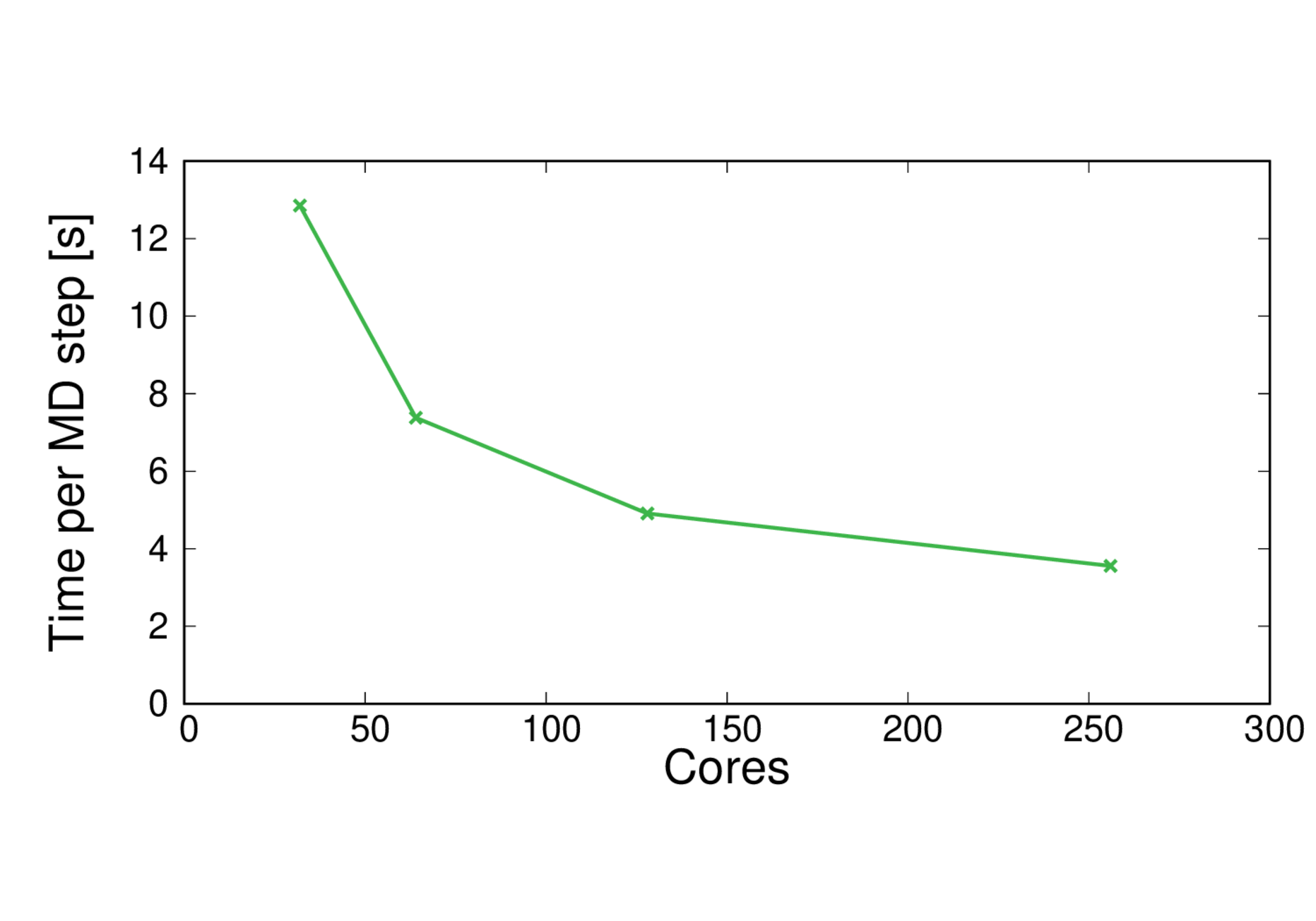
Performance@256 cores: 24 ps/day, Parallel efficiency@256 cores: 45%
MQAE-B3LYP
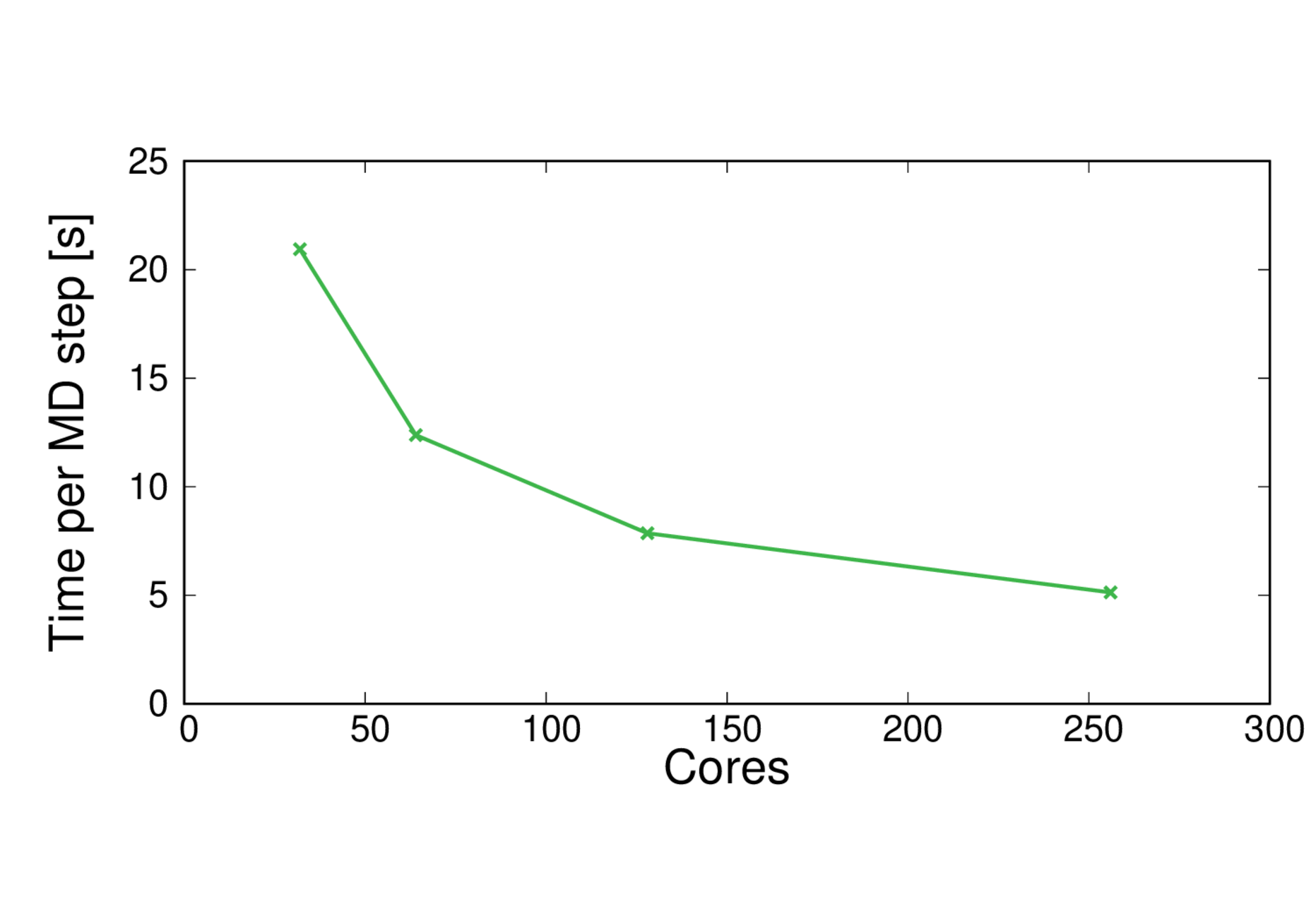
Performance@256 cores: 17 ps/day, Parallel efficiency@256 cores: 51%
MQAE-B3LYP-large
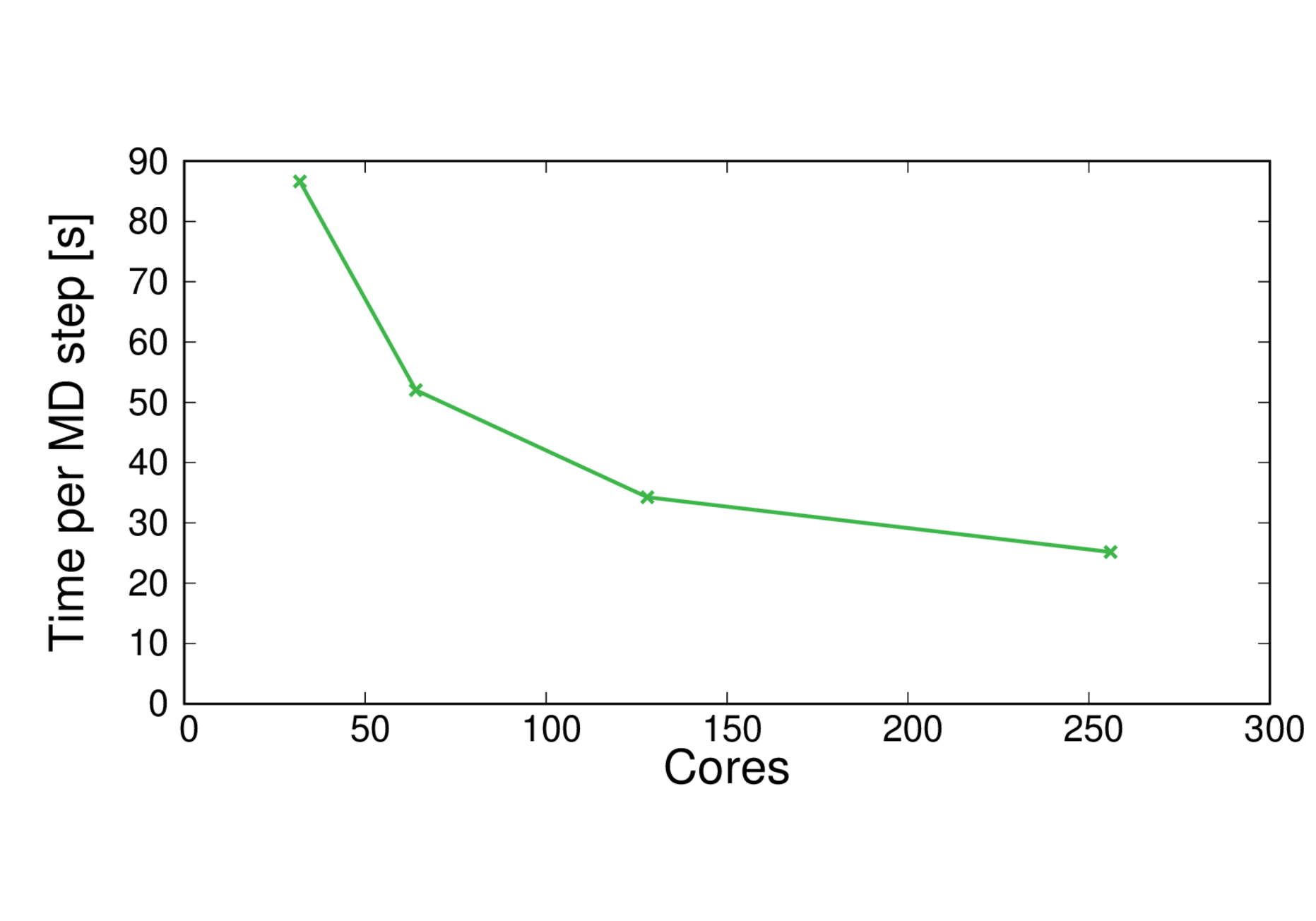
Performance@256 cores: 3.4 ps/day, Parallel efficiency@256 cores: 43%
CBD_PHY-PBE
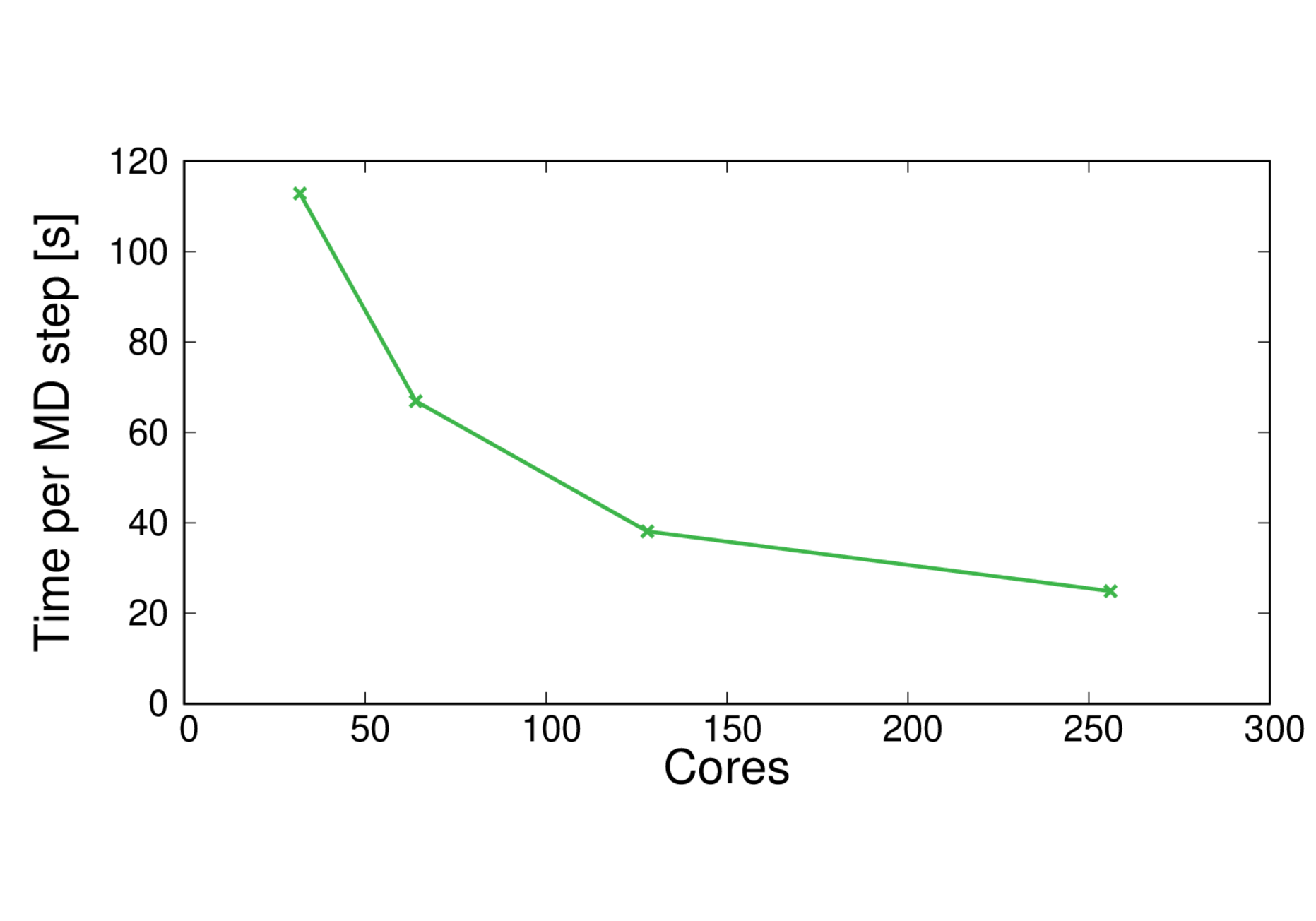
Performance@256 cores: 3.5 ps/day, Parallel efficiency@256 cores: 56%
CBD_PHY-PBE0
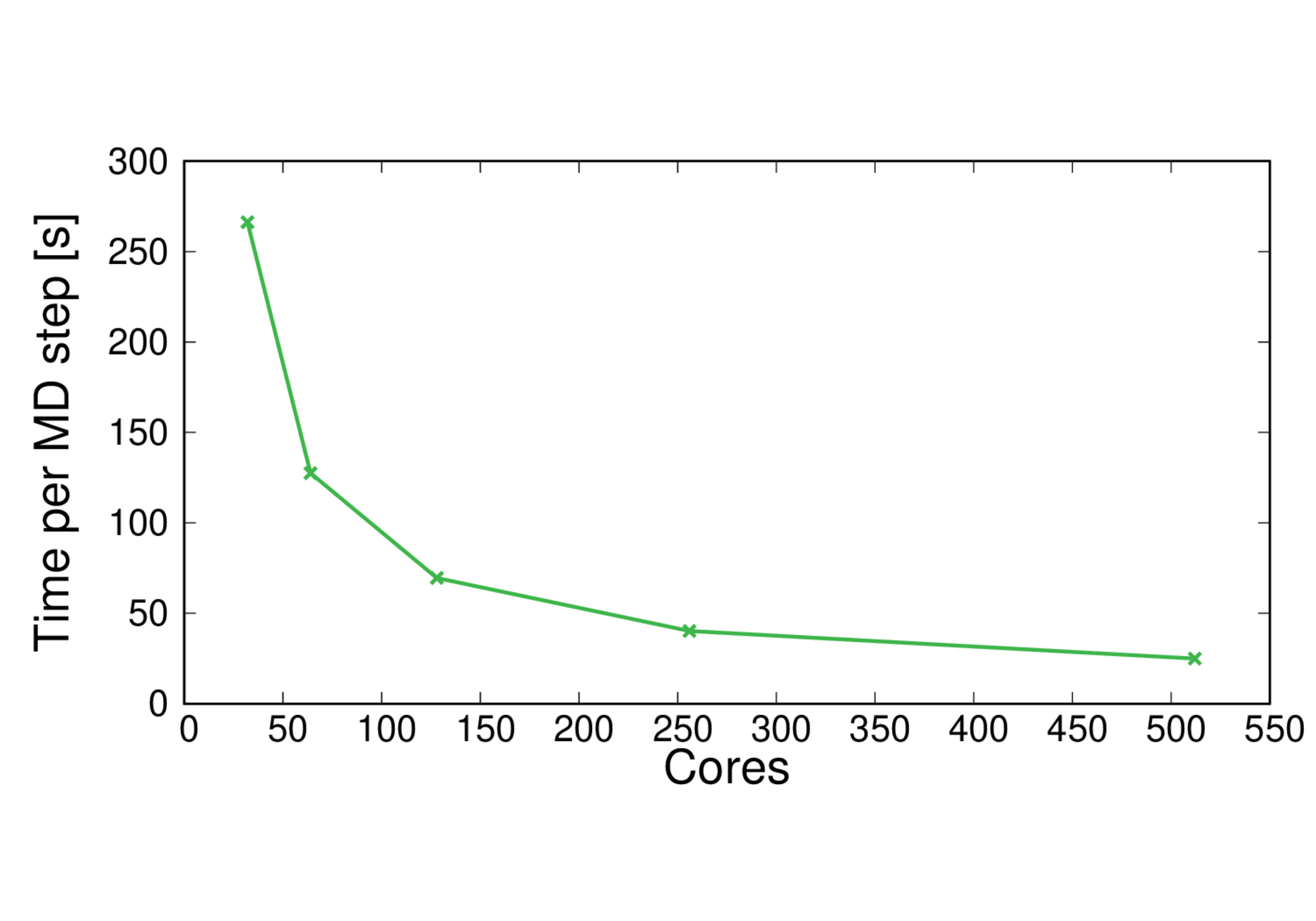
Performance@256 cores: 2.1 ps/day, Parallel efficiency@256 cores: 83%
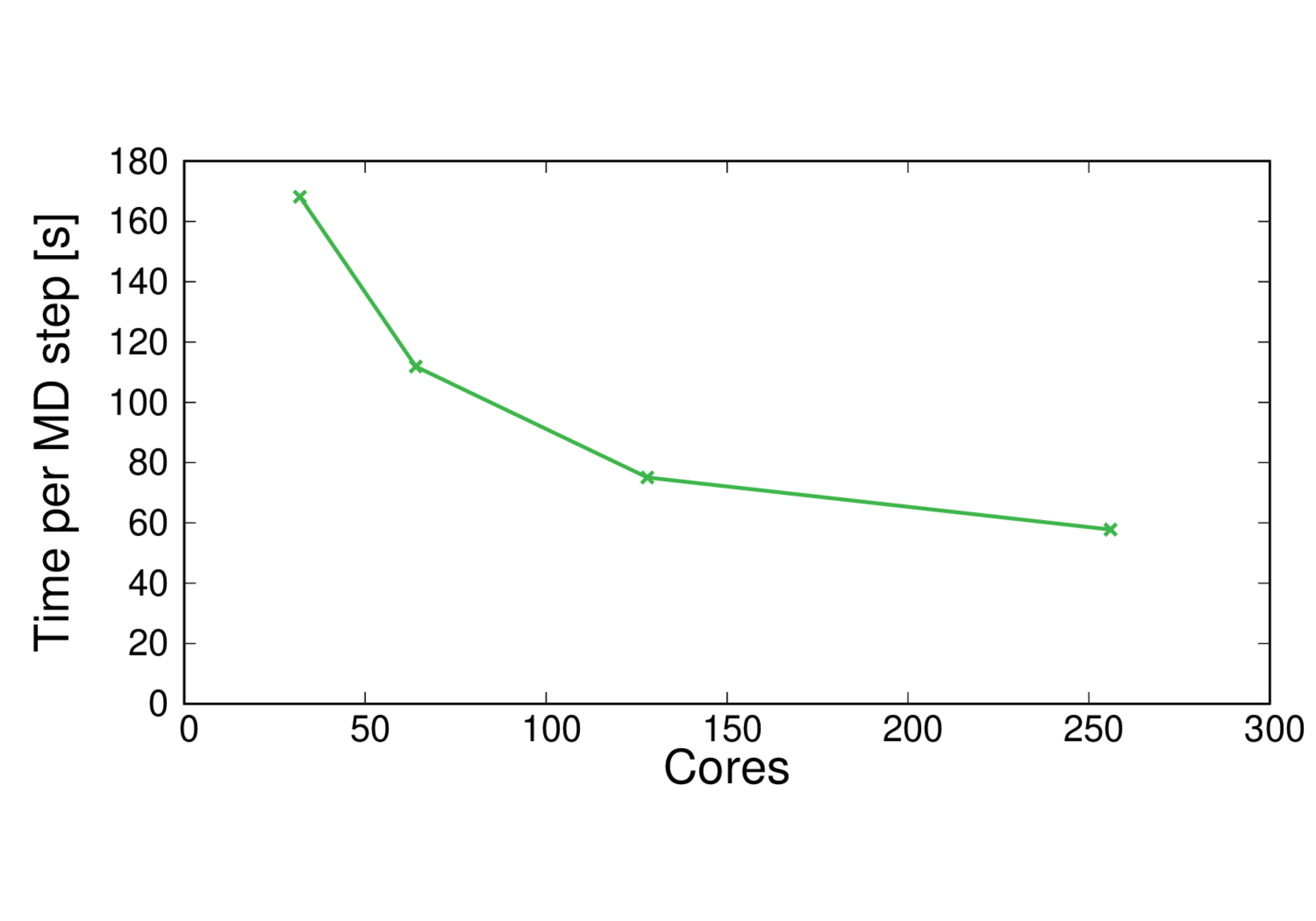
ClC-19-BLYP
Performance@128 cores: 5.7 ps/day, Parallel efficiency@128 cores: 58%
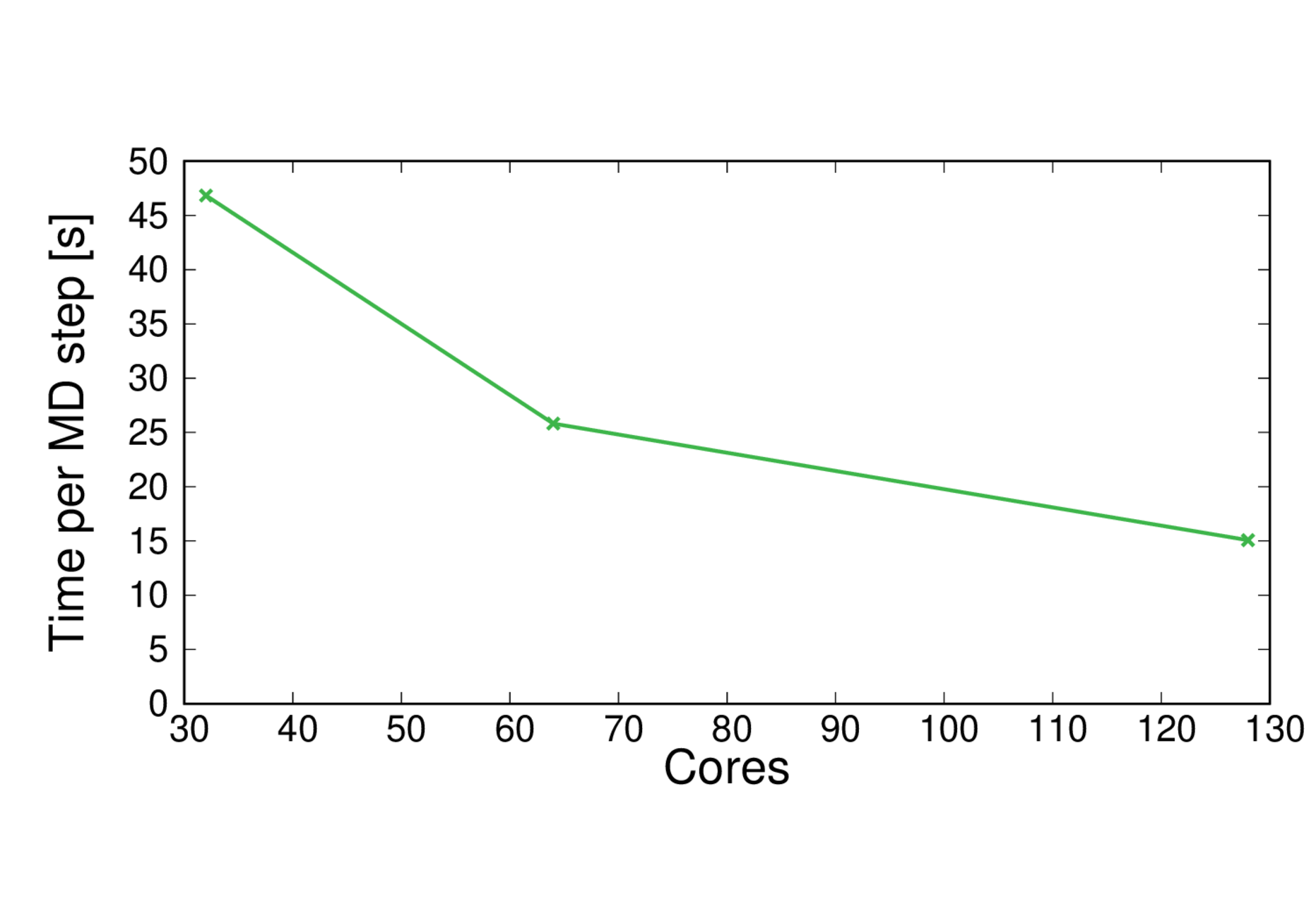
ClC-253-BLYP
Performance@256 cores: 1.5 ps/day, Parallel efficiency@256 cores: 21%
GPU benchmark results
All results are reported for Cirrus. The Cirrus GPU nodes contain 4 GPUs per node and 20 CPU cores.
MQAE-BLYP
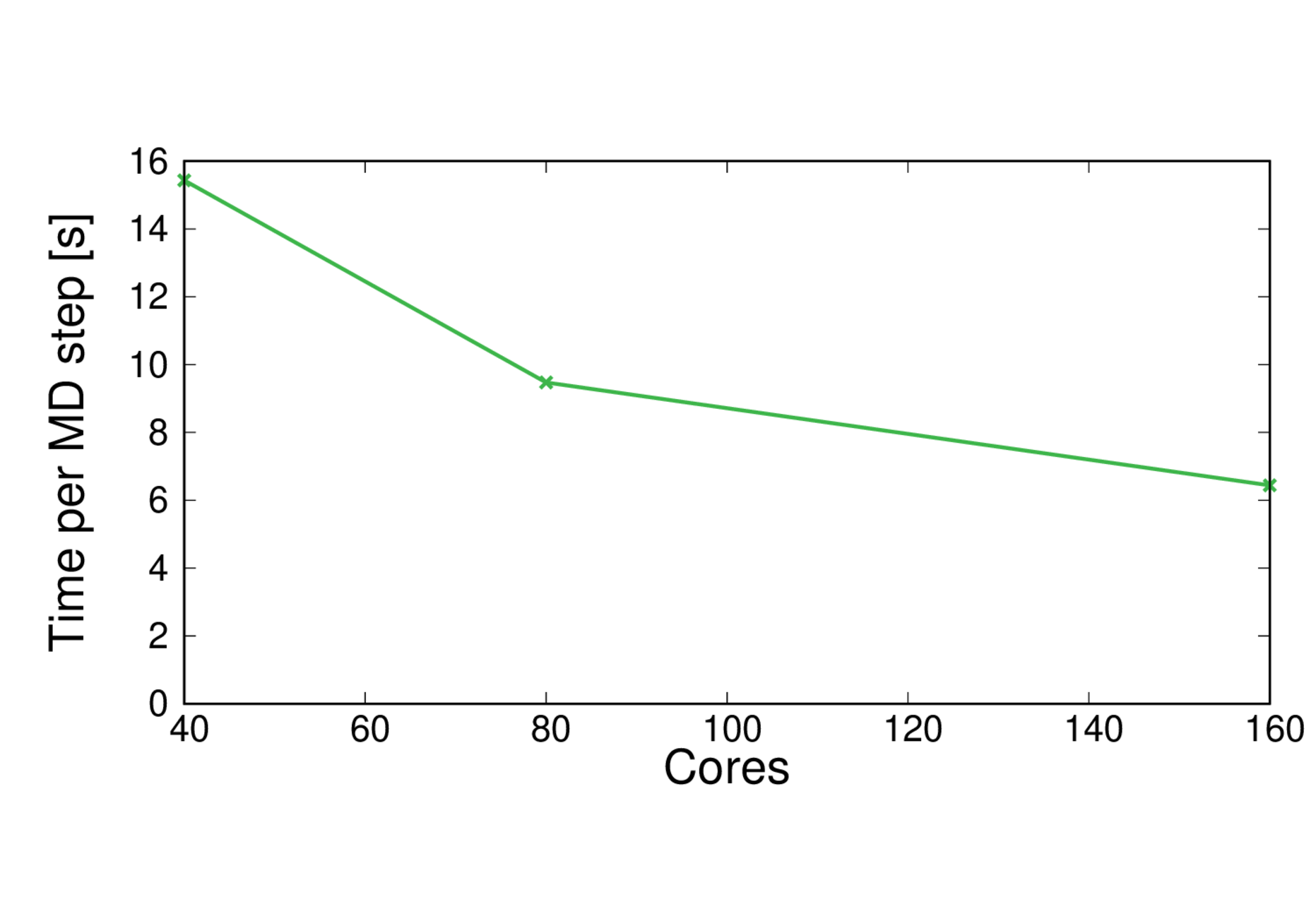
Performance@160 cores: 13.4 ps/day, Parallel efficiency@160 cores: 60%
MQAE-B3LYP
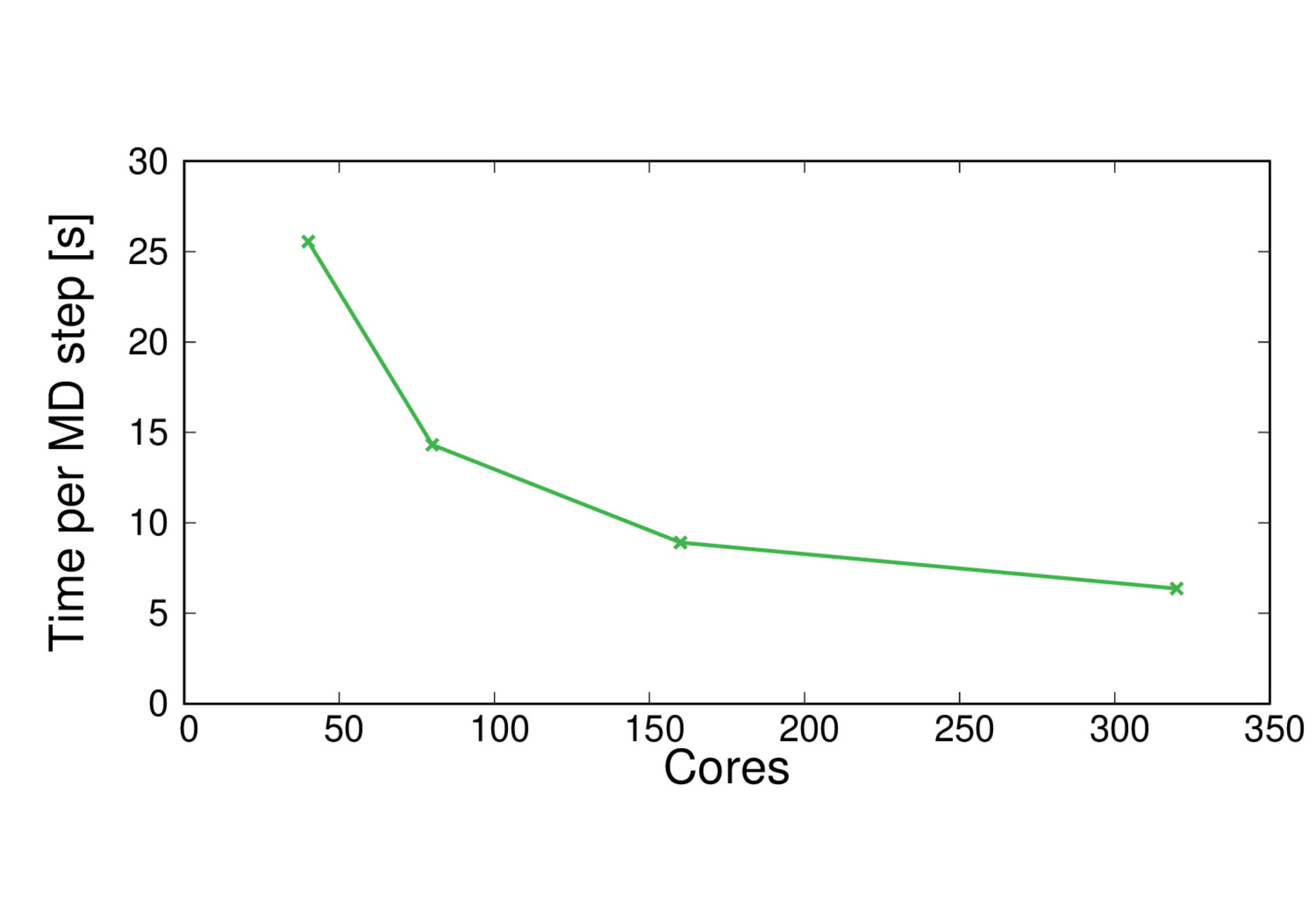
Performance@160 cores: 9.7 ps/day, Parallel efficiency@160 cores: 71%
MQAE-B3LYP-large
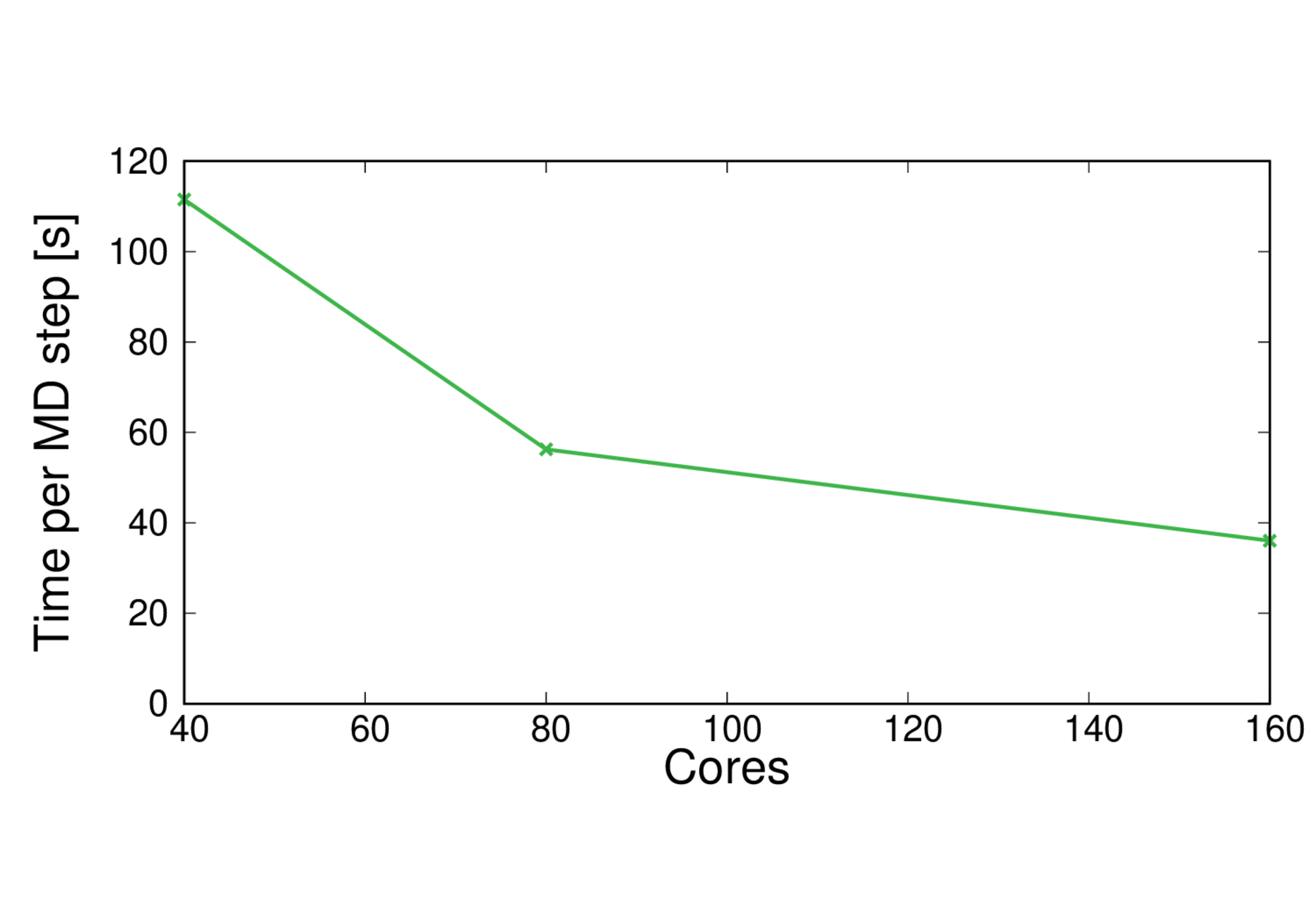
Performance@160 cores: 9.7 ps/day, Parallel efficiency@160 cores: 71%
CBD_PHY-PBE
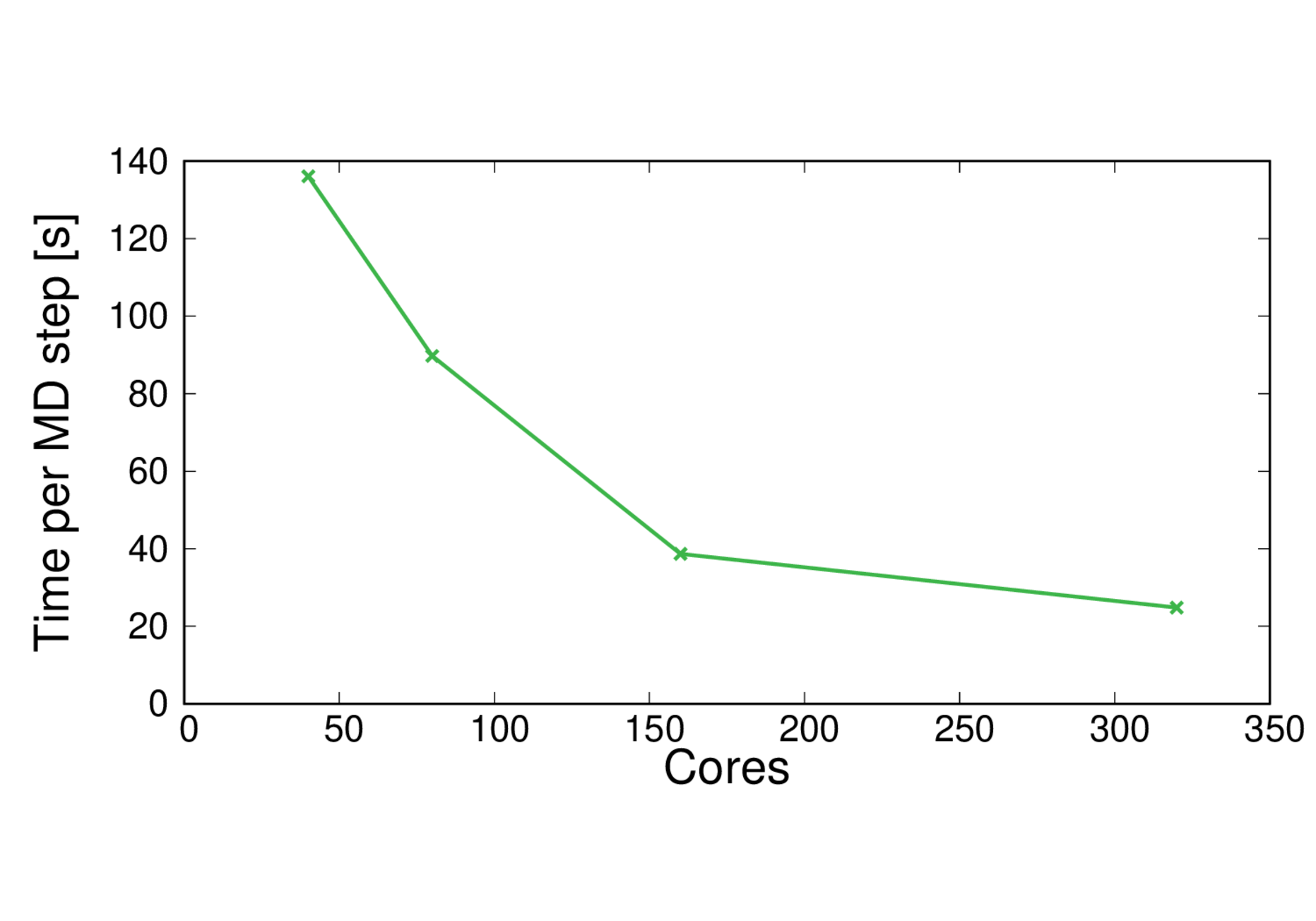
Performance@320 cores: 3.5 ps/day, Parallel efficiency@320 cores: 69%
CBD_PHY-PBE0
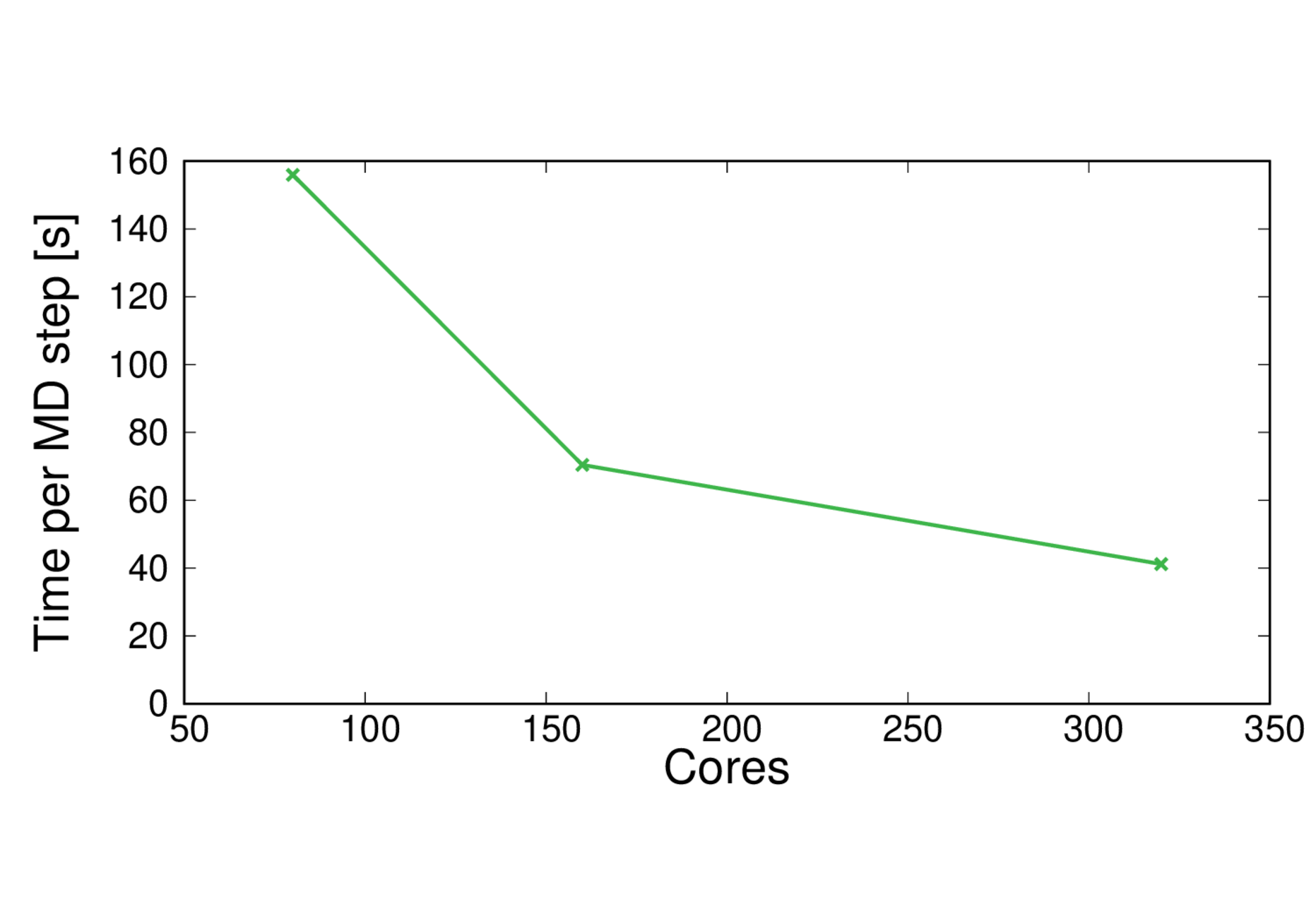
Performance@320 cores: 2.1 ps/day
ClC-19-BLYP
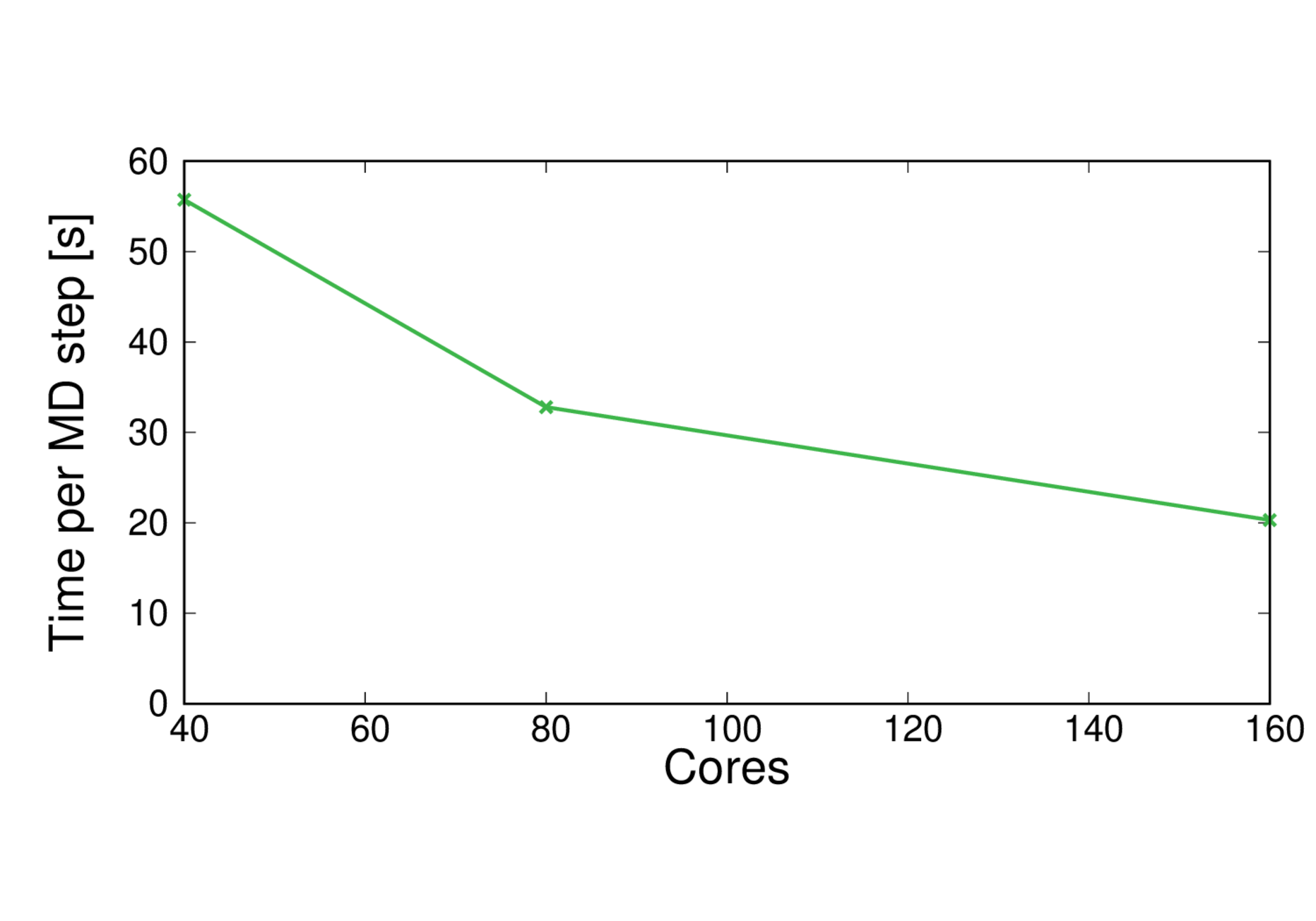
Performance@160 cores: 4.3 ps/day, Parallel efficiency@160 cores: 69%
ClC-253-BLYP
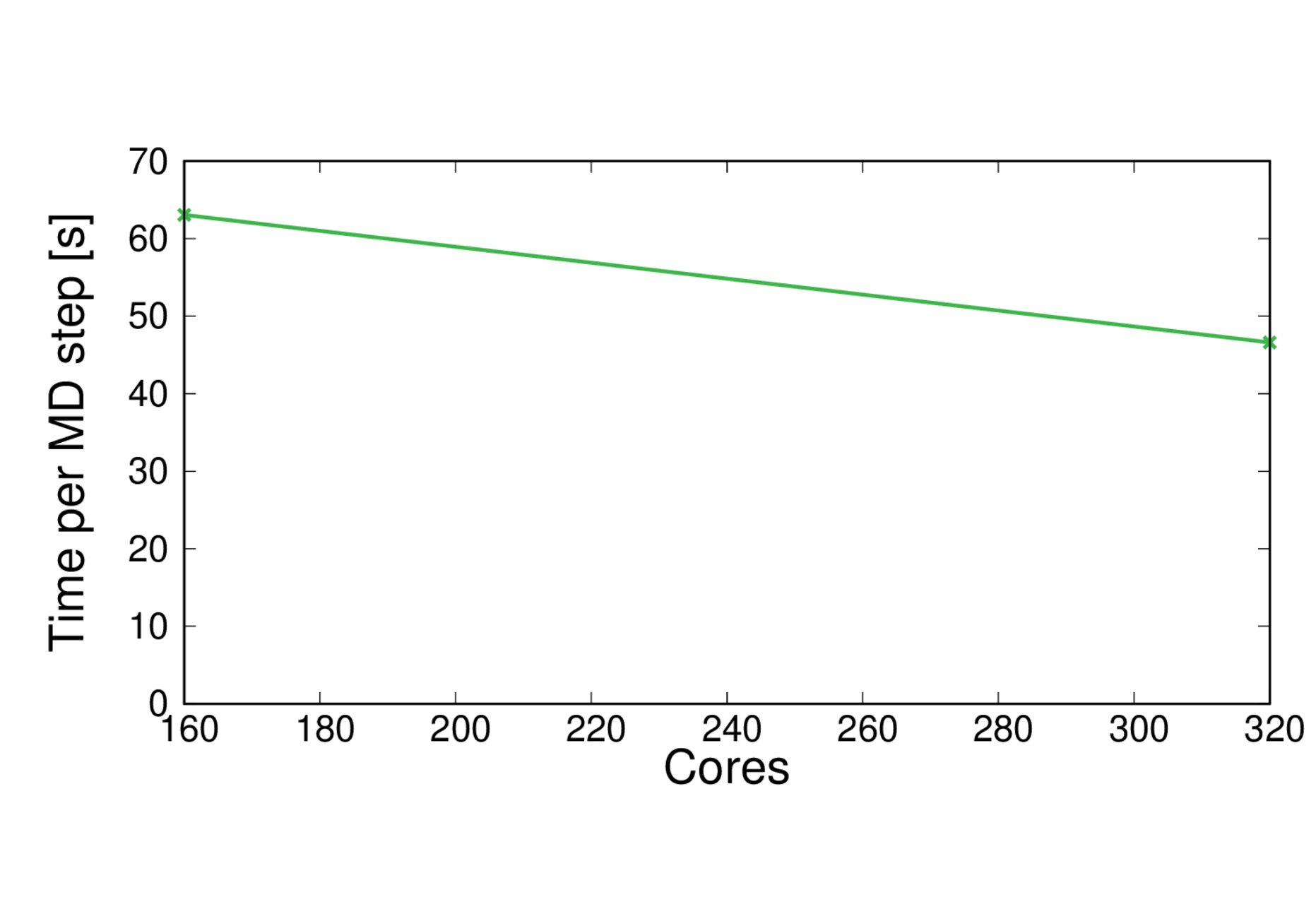
Performance@160 cores: 1.4 ps/day, Performance@320 cores: 1.9%