Practical: GROMACS + CP2K Part II
Overview
Teaching: 15 min
Exercises: 45 minQuestions
How to setup a simple QMMM system in the GROMACS-CP2K interface?
How to setup protein simulations with QMMM, breaking covalent bonds?
Objectives
Learning how to treat QM-MM interactions in fully periodic system
Perform simulation of the simple QM system in MM water
Setup protein system using default interface parameters
Preparing for the tutorial
Everything, which is written inside the gray box are a commands, that should be executed in the terminal window, string-by-string, each following with the ENTER button.
Please note that <...> in the commands means, that everything, including <> symbols, must be replaced with your own specific information. Be careful!
Helpful utilities and commands
Some exercises will require usage of
lessLinux tool for looking up into the content of files. In case you are not familiar with it, here is a short list of hotkeys, which could be used inside LESS editor:
- q – exit
- / – search for a pattern which will take you to the next occurrence
- n – for next match in forward
- N (SHIFT+n) – for previous match in backward
- g – go to the start of file
- G (SHIFT+g) – go to the end of file
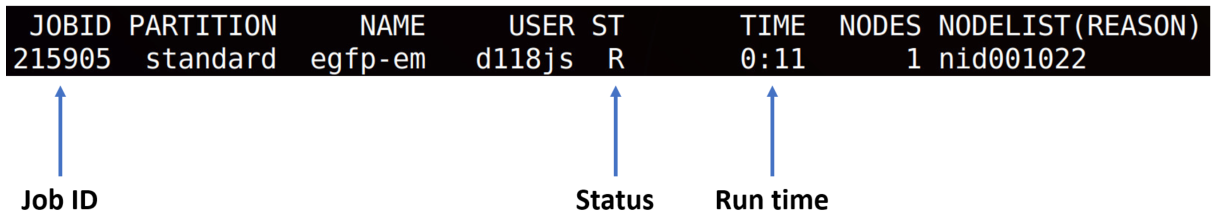
All exercises will require you to submit job for computing using
sbatch run.shcommand. To check status of your job following commands would be useful.
squeue -u <your login name>- checks status of all your jobs. Output will look like that:scancel <JOBID>will remove the job, if you occasionally submitted it.
Setting up tutorial environment
Let’s start the tutorial with the following steps
- Execute commands in the terminal:
module load gromacs-cp2k
cd /work/ta025/ta025/<your login name> - And go to the tutorial directory
cd tutorial
Exercise 3: Setting up simple QM system in MM water box
1) Go to stilbene_water directory:
cd stilbene_water
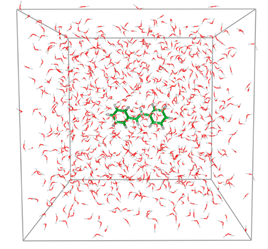
In the directory located forcefield, topology and stilbene-sol.pdb file with a geometry of stilbene in the water box
You can download it and inspect structure with VMD or PyMOL.
2) Generate QMMM index file:
gmx_cp2k make_ndx -f stilbene-sol.pdb
> 2
> name 6 QMatoms
> q
File index.ndx should appear in the directory
3) Lets perform energy minimization first for that molecule using QMMM interface
Generate Gromacs-CP2K simulation file:
gmx_cp2k grompp -f em-qmmm.mdp -p topol.top -c stilbene-sol.pdb -n index.ndx -o stilbene-sol-opt.tpr
files stilbene-sol-opt.tpr, stilbene-sol-opt.inp and stilbene-sol-opt.pdb should appear in the directory
4) Run QMMM simulation:
sbatch run-em.sh
5) While job is running you can check the content of stilbene-sol-opt.inp
less stilbene-sol-opt.inp
and of em-qmmm.mdp
less em-qmmm.mdp
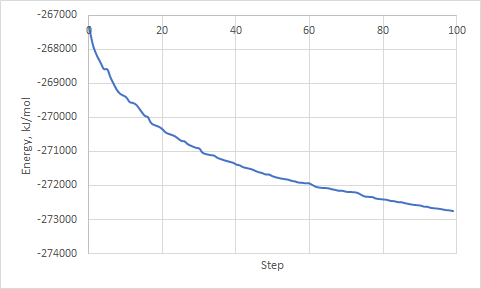
6) At the end of the job use the following command to extract potential energy:
gmx_cp2k energy
and choose 6 Potential
File energy.xvg should appear in the directory. It contains data with Potential energy (kJ/mol) against optimization step. You can open it in Grace or copy data into any other software (i.e. Excel).
You can also download trajectory file traj.trr and render it using your favorite software (e.g. VMD, PyMOL).
7) Next we will perform short (100 fs) MD simulation with QMMM. At first look into the md-qmmm-nvt.mdp file:
less md-qmmm-nvt.mdp
It contains parameters for performing dynamics with periodic QMMM forces in the NVT ensemble at 300K
8) Generate Gromacs-CP2K simulation file:
gmx_cp2k grompp -f md-qmmm-nvt.mdp -p topol.top -c stilbene-sol.pdb -n index.ndx -o stilbene-sol-nvt.tpr
files stilbene-sol-nvt.tpr, stilbene-sol-nvt.inp and stilbene-sol-nvt.pdb should appear in the directory
9) Run QMMM simulation:
sbatch run-nvt.sh
10) At the end of the simulation you can download trajectory file traj.trr and render it using your favorite software (e.g. VMD, PyMOL).
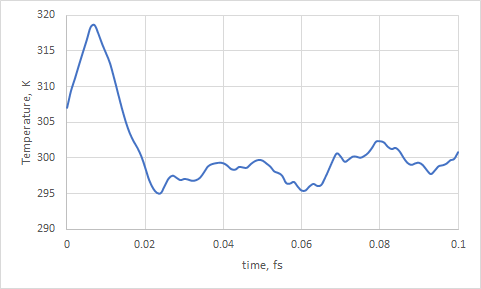
Also you could check temperature as a function of time with the following command:
gmx_cp2k energy
and choose 10 Temperature
File energy.xvg will contain data about Temperature (K) against simulation time (ps).
Notice, how temperature equilibrates and fluctuates around 300K.
Exercise 4: Large protein system setup with QMMM
1) Go to phytochrome directory:
cd ../phytochrome
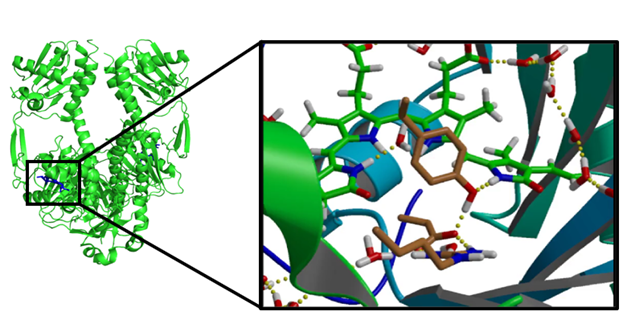
In the directory located forcefield, topology and phytochrome-sol.pdb file with a geometry of solvated phytochrome protein
You can download it and inspect structure with VMD or PyMOL.
2) Generate Gromacs-CP2K simulation file:
gmx_cp2k grompp -f md-qmmm.mdp -p topol.top -c phytochrome-sol.pdb -n index.ndx -o phytochrome.tpr
Files phytochrome.tpr, phytochrome.inp and phytochrome.pdb should appear in the directory
3) Run QMMM simulation:
sbatch run.sh
4) While job is running you can check the content of phytochrome.inp
less phytochrome.inp
and of md-qmmm.mdp
less md-qmmm.mdp
5) Notice how CP2K input file has changed. Four &LINK sections appeared at the end of &QMMM part. They are notifying CP2K that broken chemical bond should be treated between MM and QM atoms. From the side of QM part that that result in adding “fake” Hydrogen Link-atom between QM and MM atoms. This is automatically done inside the CP2K.
6) You could also look up in the index.ndx which atoms are marked up as QMatoms group
Try to render QMatoms in VMD or PyMOL. They are also marked up as residue QM in phytochrome.pdb file generated during gmx_cp2k grompp
Key Points
GROMACS-CP2K allows you to perform fully periodic QMMM simulations
Interface will take care of QMMM topology modifications, QM box dimensions and provide initial parameters
Broken covalent bonds will be treated automatically